Authentic TF-SHIV Model of HIV-1 Immunopathogenesis
In recent years, we have generated a body of work showing that TF-SHIVs reproduce features of HIV-1 Viral dynamics, pathogenesis, immune responses, persistence, and viral rebound. The TF SHIV-rhesus macaque (RM) model system was born of our discovery that substitution of a single critical amino acid residue at Env position 375 in the CD4 binding pocket of HIV-1 gp120 enhances the binding of HIV-1 Envs to rhesus CD4, thereby enabling persistent replication in RMs [ref]. Importantly, this substitution confers efficient entry and replication by every tested Env, but does not change their antigenic profile, tier 2 neutralization status, or conformational state. Using this approach, we generated SHIVs containing clade A, B, C, D, E and CRF TF Envs, each encoding the TF HIV-1 Env modified only at Env375 with a bulky, hydrophobic residue selected in vivo in multiple outbred RM (Fig1). Each TF SHIV demonstrated excellent early viral kinetics and pathogenesis that mirrored HIV-1, with depletion of CD4 T cells, lymphoid fibrosis, and clinical progression to simian AIDS [Ref]. While some TF SHIVs caused lower viral load set points or frequent spontaneous control, a SHIV encoding a clade D TF Env from participant 191859 (SHlV.D), was consistently one of the best replicators. Recently, we expanded this model to incorporate genetic barcoding with virus inocula containing mixtures of bnAb (broadly neutralizing antibodies)-sensitive and resistant viruses to create a molecularly defined virus model. In this system, each animal is infected with a precise ratio of TF SHIVs encoding wildtype (WT) and escape mutant (EM) viruses, which have similar replication kinetics but markedly different sensitivities to V3-glycan bnAbs.
This Novel NHP model is currently used in our laboratory to dissect the effects of bnAbs administration at ART initiation to characterize the baseline reservoir formation, the global impact of bnABs, and the specific role of effector function. This strategy allows us to test:
if the bnABs at ART initiation decrease the size, diversity, and persistence of the reservoir
Increase the potency and durability of humoral and cellular anti-HIV immune responses
Decrease virus activation rates with boosted immune responses at treatment interruption
Specifically, our lab is interested in characterizing:
The impacts of bnAbs at ART initiation on HIV-specific immune responses
The impact of bnAbs at ART initiation on virus reactivation at ATI
Understanding the biology and impacts of bnAbs on the HIV-1 immune responses and the effect on the reservoir would have substantial significance to the HIV cure field, as will elucidate mechanisms of reservoir formation, characterize immunomodulation of anti-HIV immune responses, dissect the roles of antibody neutralization and effector functions, and inform our understanding of promising cure interventions.
Characterization of the effects of bnAbs at ART initiation
The potential of monoclonal antibodies
The use of monoclonal antibodies (mAbs) for HIV-1 holds great promise, but HIV-1 envelope trimer is a challenging target for humoral immunity, as it is highly glycosylated, masks its conserved regions, and is tolerant of variation in exposed regions. During infection neutralizing antibody responses drive virus selection, but lag behind rapidly adapting contemporaneous virus populations. In rare individuals, these successive waves of virus evolution and antibody responses lead to the development of unique broadly neutralizing antibodies (bNAbs) that successfully navigate the glycosylated, variable Env to bind conserved regions. While ineffective at virus suppression in the hosts in which they evolved, their use prior to infection or in people living with HIV (PWH) with sensitive virus holds promise. Advances in B cell biology and molecular virology have enabled the discovery, characterization, and commercial development of these rare bNAbs, which now drive many strategies for the prevention, treatment, and cure of HIV. Yet, virus resistance remains the central vulnerability of effective bNAb use. Indeed, efficacy in recent trials for prevention and virus suppression was limited by resistance.
Effective strategies to combat HIV-1 via bNAbs require a better understanding of the selective pressure imposed by bNAbs and novel approaches to limit virus escape.
Our laboratory, based on a multi-disciplinary team, has secured samples or virus sequences from recent or ongoing prevention and treatment studies of VRC01-class CD4 binding site (CD4bs)-targeting monotherapy, as well as combination therapy with V3-glycan-targeting. By using these unique samples our lab aims to:
map the in vivo escape pathways of virus replicating in the presence of sub-suppressive levels of this clinically relevant bNAbs
Using these escape variants, to identify putative complementary bNAbs with maintained or inverse antibody sensitivities from rationally designed panels of candidate bNAbs
Characterize the autologous neutralizing antibody (anAb) response in the treatment cohorts, to determine the capacity of anAbs to impede virus escape from administered bNAbs
Test the most promising complementary bNAbs to restrict virus escape in vivo in a validated barcoded TF SHIV/NHP model
Our overall objective is to elucidate in vivo mapping of virus escape and, by using an authentic NHP model, to elucidate basic mechanisms of virus resistance to bNAbs and inform more effective use of bNAbs across the HIV prevention, treatment, and cure fields.
CD4bs bNAb variability. Angles of approach for CD4bs antibodies on the trimer spike (gray) in pre-fusion near-native conformation colored by subgroup. Binding orientations of CD4 (yellow), CDR H3-dominated bNAbs (red), V- gene restricted bnAbs are indicated with lines while N-linked glycans (teal) and the CD4-bs (yellow surface) are also shown.
Characterization of persistent myeloid reservoirs in the brain to assess the safety and efficacy of CRISPR-based eradication streategies.
Emerging evidence suggests that myeloid reservoirs in the CNS and elsewhere may compromise important and understudied sources of persistent viruses (ref). Human studies have shown that brain macrophages harbor HIV DNA and are persistently activated and dysfunctional in ART-suppressed PLWH (people living with HIV).
Our lab aims to characterize the CNC virus reservoirs that persist in long-term ART in our above-mentioned authentic NHP model. Specifically we aim to:
Define persistent CNS and systemic reservoirs in latently infected SHIV.D-infected RM
Optimize AAV-delivered CRISPR/Cas9 for barcoded SHIV.D. excision in CNS
Assess the safety and efficacy of CRISPR excision of latent SHIV.D in vivo
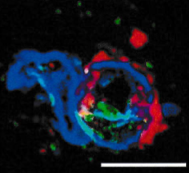
DNA and RNAscope of SHIV-infected RM brain cortex. SHIV-DNA (green), SHIV-RNA (red), nuclei (blue). Scale bar = 10 micrometer